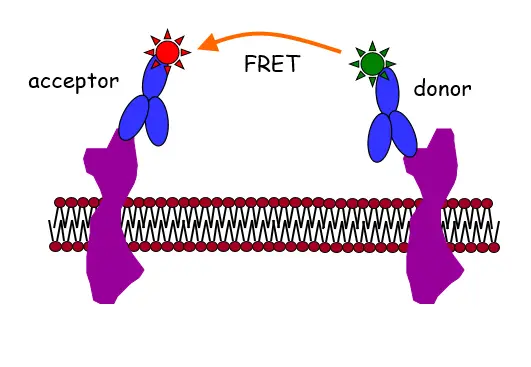
Fluorescence labeling of proteins is usually achieved by fluorescent monoclonal antibodies (see figure above) or with fluorescent protein constructs. We have achieved the following results in our method development efforts:
- We have shown that intensity-based FRET experiments carried out at high excitation intensities, commonly achieved in confocal microscopy, lead to severely underestimated FRET values if the analysis is carried out with the conventional equations disregarding fluorophore saturation. Fluorophore saturation is a situation when most fluorophores are in the excited state. The effect of this phenomenon on donor quenching is summarized in the figure below:
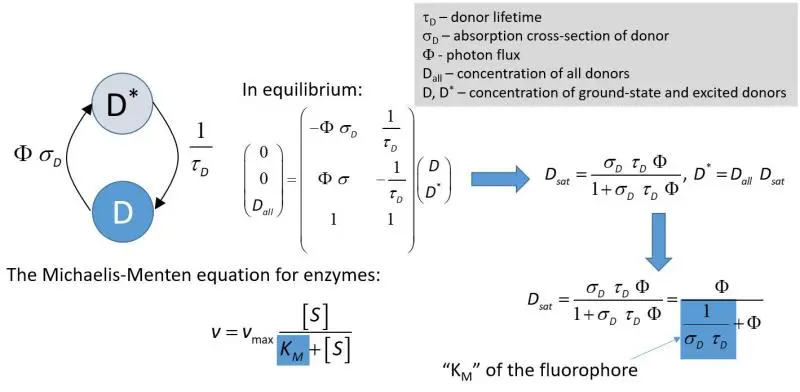
A complete treatment of this condition is available
- in this PDF presentation
- in a paper published in Anal. Chem.
- in the rFRET Matlab application
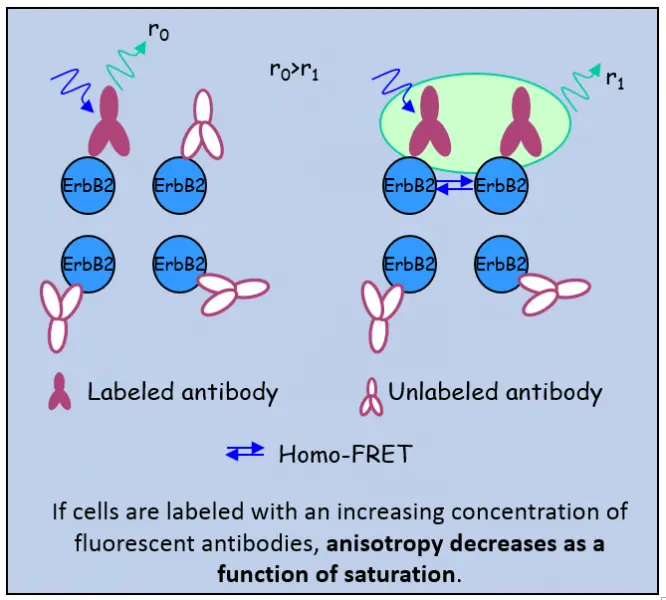
- we have developed a method for the investigation of the medium- and large-scale clustering of proteins based on flow cytometric measurement of homo-FRET
- we have developed a method for the accurate estimation of the FRET efficiency in the presence of poor signal-to-noise ratio based on maximum likelihood estimation
- we have proven using simulations that the mean of pixelwise FRET efficiencies calculated from microscopic measurements in the presence of poor signal-to-noise ratio is an unbiased estimator of the real FRET efficiency
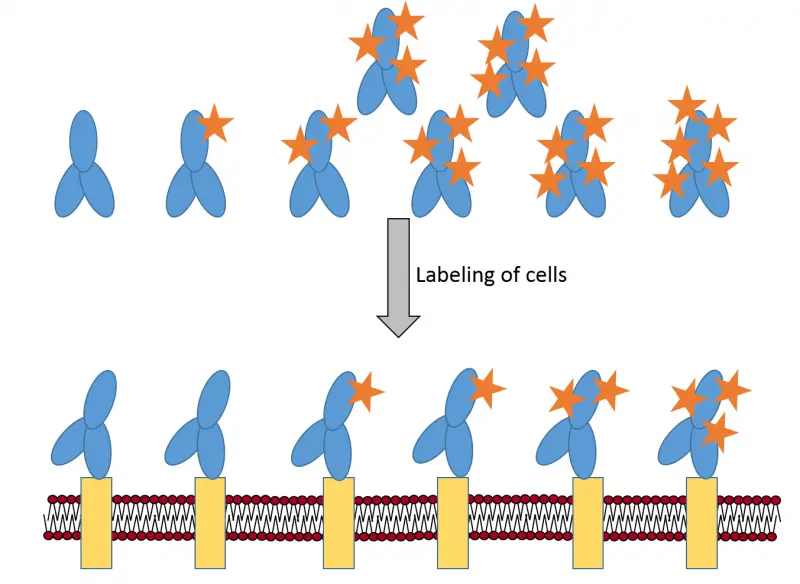
- we have proven that fluorescence labeling deteriorates antibody affinity and consequently the average degree of labeling (DOL) of the cell-bound antibody fraction is lower than that of the stock.
In our current research efforts we would like to elucidate
- whether the amino groups of certain amino acids of antibodies are preferentially labeled in the conjugation reaction using mass spectrometry